Some of you may have heard of ‘Folding@Home’ from it’s recent publicity with Playstation 3 and the release of new ATI graphics cards. If you haven’t; Folding@Home is a project started by Stanford university to better understand the process of protein folding. To help research into what proteins do, a distributed computing project has been set up to simulate this cycle.
Due to the fact so much processing power is required, the project is distributed across normal home computers opposed to only running on super computers. This allows maximum processing power. Anyone at home that wants to participate can download the free software application from here. With so many participants using their home PCs to fold, the combined processing speed is greater than the world’s fastest super computer.
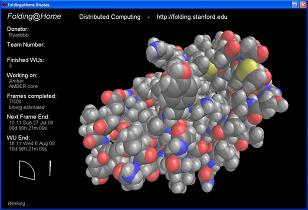
The PC version utilizes the CPU to perform simulations of folding. To do this, the program automatically downloads a small file and will begin folding, frame by frame. Once you have completed a set number of frames the information will be sent back to Stanford and a new download will begin. This is all done in the background while you can be doing other things on your computer. If you like, you can set it so it only runs when your screensaver is active.
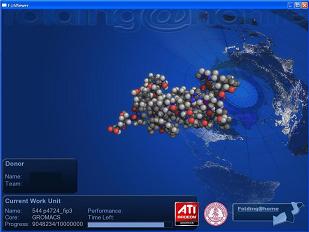
There is also a GPU version of the program that performs much faster than the CPU version as it is running on the graphics card and not restricted to the central processing unit(CPU). There is also a Playstation 3 client that folds while the game is not in use.
The protein simulations will help us to better understand the development of many diseases, including; Alzheimer’s disease sickle-cell disease (drepanocytosis), Parkinson’s disease, BSE (mad cow disease), cancer, Huntington’s disease, cystic fibrosis, osteogenesis imperfecta, alpha 1-antitrypsin deficiency, and other aggregation-related diseases.
Here are screenshots of both the CPU and GPU versions of the program running.
Click to Enlarge.
Recent Comments